 Front page > Programming > Feature Selection with the IAMB Algorithm: A Casual Dive into Machine Learning
Front page > Programming > Feature Selection with the IAMB Algorithm: A Casual Dive into Machine Learning
Feature Selection with the IAMB Algorithm: A Casual Dive into Machine Learning
So, here’s the story—I recently worked on a school assignment by Professor Zhuang involving a pretty cool algorithm called the Incremental Association Markov Blanket (IAMB). Now, I do not have a background in data science or statistics, so this is new territory for me, but I love to learn something new. The goal? Use IAMB to select features in a dataset and see how it impacts the performance of a machine-learning model.
We’ll go over the basics of the IAMB algorithm and apply it to the Pima Indians Diabetes Dataset from Jason Brownlee's datasets. This dataset tracks health data on women and includes whether they have diabetes or not. We’ll use IAMB to figure out which features (like BMI or glucose levels) matter most for predicting diabetes.
What’s the IAMB Algorithm, and Why Use It?
The IAMB algorithm is like a friend who helps you clean up a list of suspects in a mystery—it’s a feature selection method designed to pick out only the variables that truly matter for predicting your target. In this case, the target is whether someone has diabetes.
- Forward Phase: Add variables that are strongly related to the target.
- Backward Phase: Trim out the variables that don’t really help, ensuring only the most crucial ones are left.
In simpler terms, IAMB helps us avoid clutter in our dataset by selecting only the most relevant features. This is especially handy when you want to keep things simple boost model performance and speed up the training time.
Source: Algorithms for Large-Scale Markov Blanket Discovery
What’s This Alpha Thing, and Why Does it Matter?
Here’s where alpha comes in. In statistics, alpha (α) is the threshold we set to decide what counts as "statistically significant." As part of the instructions given by the professor, I used an alpha of 0.05, meaning I only want to keep features that have less than a 5% chance of being randomly associated with the target variable. So, if a feature’s p-value is less than 0.05, it means there’s a strong, statistically significant association with our target.
By using this alpha threshold, we’re focusing only on the most meaningful variables, ignoring any that don’t pass our “significance” test. It’s like a filter that keeps the most relevant features and tosses out the noise.
Getting Hands-On: Using IAMB on the Pima Indians Diabetes Dataset
Here's the setup: the Pima Indians Diabetes Dataset has health features (blood pressure, age, insulin levels, etc.) and our target, Outcome (whether someone has diabetes).
First, we load the data and check it out:
import pandas as pd # Load and preview the dataset url = 'https://raw.githubusercontent.com/jbrownlee/Datasets/master/pima-indians-diabetes.data.csv' column_names = ['Pregnancies', 'Glucose', 'BloodPressure', 'SkinThickness', 'Insulin', 'BMI', 'DiabetesPedigreeFunction', 'Age', 'Outcome'] data = pd.read_csv(url, names=column_names) print(data.head())
Implementing IAMB with Alpha = 0.05
Here’s our updated version of the IAMB algorithm. We’re using p-values to decide which features to keep, so only those with p-values less than our alpha (0.05) are selected.
import pingouin as pg
def iamb(target, data, alpha=0.05):
markov_blanket = set()
# Forward Phase: Add features with a p-value alpha
for feature in list(markov_blanket):
reduced_mb = markov_blanket - {feature}
result = pg.partial_corr(data=data, x=feature, y=target, covar=reduced_mb)
p_value = result.at[0, 'p-val']
if p_value > alpha:
markov_blanket.remove(feature)
return list(markov_blanket)
# Apply the updated IAMB function on the Pima dataset
selected_features = iamb('Outcome', data, alpha=0.05)
print("Selected Features:", selected_features)
When I ran this, it gave me a refined list of features that IAMB thought were most closely related to diabetes outcomes. This list helps narrow down the variables we need for building our model.
Selected Features: ['BMI', 'DiabetesPedigreeFunction', 'Pregnancies', 'Glucose']
Testing the Impact of IAMB-Selected Features on Model Performance
Once we have our selected features, the real test compares model performance with all features versus IAMB-selected features. For this, I went with a simple Gaussian Naive Bayes model because it’s straightforward and does well with probabilities (which ties in with the whole Bayesian vibe).
Here’s the code to train and test the model:
from sklearn.model_selection import train_test_split
from sklearn.naive_bayes import GaussianNB
from sklearn.metrics import accuracy_score, f1_score, roc_auc_score
# Split data
X = data.drop('Outcome', axis=1)
y = data['Outcome']
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=42)
# Model with All Features
model_all = GaussianNB()
model_all.fit(X_train, y_train)
y_pred_all = model_all.predict(X_test)
# Model with IAMB-Selected Features
X_train_selected = X_train[selected_features]
X_test_selected = X_test[selected_features]
model_iamb = GaussianNB()
model_iamb.fit(X_train_selected, y_train)
y_pred_iamb = model_iamb.predict(X_test_selected)
# Evaluate models
results = {
'Model': ['All Features', 'IAMB-Selected Features'],
'Accuracy': [accuracy_score(y_test, y_pred_all), accuracy_score(y_test, y_pred_iamb)],
'F1 Score': [f1_score(y_test, y_pred_all, average='weighted'), f1_score(y_test, y_pred_iamb, average='weighted')],
'AUC-ROC': [roc_auc_score(y_test, y_pred_all), roc_auc_score(y_test, y_pred_iamb)]
}
results_df = pd.DataFrame(results)
display(results_df)
Results
Here’s what the comparison looks like:
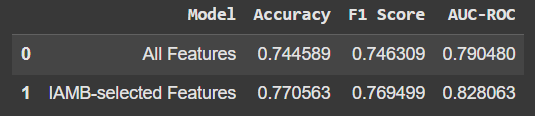
Using only the IAMB-selected features gave a slight boost in accuracy and other metrics. It’s not a huge jump, but the fact that we’re getting better performance with fewer features is promising. Plus, it means our model isn’t relying on “noise” or irrelevant data.
Key Takeaways
- IAMB is great for feature selection: It helps clean up our dataset by focusing only on what really matters for predicting our target.
- Less is often more: Sometimes, fewer features give us better results, as we saw here with a small boost in model accuracy.
- Learning and experimenting is the fun part: Even without a deep background in data science, diving into projects like this opens up new ways to understand data and machine learning.
I hope this gives a friendly intro to IAMB! If you’re curious, give it a shot—it’s a handy tool in the machine learning toolbox, and you might just see some cool improvements in your own projects.
Source: Algorithms for Large-Scale Markov Blanket Discovery
-
 Beyond `if` Statements: Where Else Can a Type with an Explicit `bool` Conversion Be Used Without Casting?Contextual Conversion to bool Allowed Without a CastYour class defines an explicit conversion to bool, enabling you to use its instance 't' di...Programming Published on 2024-11-18
Beyond `if` Statements: Where Else Can a Type with an Explicit `bool` Conversion Be Used Without Casting?Contextual Conversion to bool Allowed Without a CastYour class defines an explicit conversion to bool, enabling you to use its instance 't' di...Programming Published on 2024-11-18 -
 Can You Have Multiple Classes in a Single Java File?Multiple Classes in a Java FileIn Java, it is possible to have multiple classes within a single .java file. However, there can only be one public top-...Programming Published on 2024-11-18
Can You Have Multiple Classes in a Single Java File?Multiple Classes in a Java FileIn Java, it is possible to have multiple classes within a single .java file. However, there can only be one public top-...Programming Published on 2024-11-18 -
 What Happened to Column Offsetting in Bootstrap 4 Beta?Bootstrap 4 Beta: The Removal and Restoration of Column OffsettingBootstrap 4, in its Beta 1 release, introduced significant changes to the way column...Programming Published on 2024-11-18
What Happened to Column Offsetting in Bootstrap 4 Beta?Bootstrap 4 Beta: The Removal and Restoration of Column OffsettingBootstrap 4, in its Beta 1 release, introduced significant changes to the way column...Programming Published on 2024-11-18 -
 How do I combine two associative arrays in PHP while preserving unique IDs and handling duplicate names?Combining Associative Arrays in PHPIn PHP, combining two associative arrays into a single array is a common task. Consider the following request:Descr...Programming Published on 2024-11-18
How do I combine two associative arrays in PHP while preserving unique IDs and handling duplicate names?Combining Associative Arrays in PHPIn PHP, combining two associative arrays into a single array is a common task. Consider the following request:Descr...Programming Published on 2024-11-18 -
 How to Test PDO Database Connections and Handle Errors Effectively?Testing PDO Database ConnectionsWhen developing database installations, it is crucial to ensure the validity of database connections. This becomes par...Programming Published on 2024-11-18
How to Test PDO Database Connections and Handle Errors Effectively?Testing PDO Database ConnectionsWhen developing database installations, it is crucial to ensure the validity of database connections. This becomes par...Programming Published on 2024-11-18 -
 Does MySQL Update Queries Overwrite Existing Values When They Are The Same?MySQL Update Queries: Overwriting Existing ValuesIn MySQL, when updating a table, it's possible to encounter a scenario where the new value you sp...Programming Published on 2024-11-18
Does MySQL Update Queries Overwrite Existing Values When They Are The Same?MySQL Update Queries: Overwriting Existing ValuesIn MySQL, when updating a table, it's possible to encounter a scenario where the new value you sp...Programming Published on 2024-11-18 -
 Why does `std::atomic`\'s store use XCHG for sequential consistency on x86?Why std::atomic's store employs XCHG for sequential consistencyIn the context of std::atomic for x86 and x86_64 architectures, a store operation w...Programming Published on 2024-11-18
Why does `std::atomic`\'s store use XCHG for sequential consistency on x86?Why std::atomic's store employs XCHG for sequential consistencyIn the context of std::atomic for x86 and x86_64 architectures, a store operation w...Programming Published on 2024-11-18 -
 Why doesn't C++ directly support returning arrays from functions?Why C Disapproves of Array-Returning FunctionsThe C LandscapeIn contrast to languages like Java, C doesn't offer direct support for function...Programming Published on 2024-11-18
Why doesn't C++ directly support returning arrays from functions?Why C Disapproves of Array-Returning FunctionsThe C LandscapeIn contrast to languages like Java, C doesn't offer direct support for function...Programming Published on 2024-11-18 -
 Okay, here are some titles that would fit the content of the article: * How to Fix the \"-lGL: not found\" Error in Qt * Qt Compilation Error: \"-lGL: not found\" - What to Do * Troubleshooting \"-lGL: not found\" Error in QResolving "-lGL: not found" Error in QtWhen trying to compile a newly created project in QtCreator, some users may encounter the error "...Programming Published on 2024-11-18
Okay, here are some titles that would fit the content of the article: * How to Fix the \"-lGL: not found\" Error in Qt * Qt Compilation Error: \"-lGL: not found\" - What to Do * Troubleshooting \"-lGL: not found\" Error in QResolving "-lGL: not found" Error in QtWhen trying to compile a newly created project in QtCreator, some users may encounter the error "...Programming Published on 2024-11-18 -
 Is PHP's `eval` Function Ever Safe to Use?When (if Ever) is eval NOT Evil?While PHP's eval function has often been discouraged, its utility in PHP 5.3 is debatable. Despite the emergence o...Programming Published on 2024-11-18
Is PHP's `eval` Function Ever Safe to Use?When (if Ever) is eval NOT Evil?While PHP's eval function has often been discouraged, its utility in PHP 5.3 is debatable. Despite the emergence o...Programming Published on 2024-11-18 -
 How to Fix \"ImproperlyConfigured: Error loading MySQLdb module\" in Django on macOS?MySQL Improperly Configured: The Problem with Relative PathsWhen running python manage.py runserver in Django, you may encounter the following error:I...Programming Published on 2024-11-18
How to Fix \"ImproperlyConfigured: Error loading MySQLdb module\" in Django on macOS?MySQL Improperly Configured: The Problem with Relative PathsWhen running python manage.py runserver in Django, you may encounter the following error:I...Programming Published on 2024-11-18 -
 How to Unmarshal XML with Dynamic Attributes in Go?Golang: Unmarshalling XML with Dynamic AttributesIntroduction:In Go, encoding/xml provides an efficient and versatile way to handle XML data. However,...Programming Published on 2024-11-18
How to Unmarshal XML with Dynamic Attributes in Go?Golang: Unmarshalling XML with Dynamic AttributesIntroduction:In Go, encoding/xml provides an efficient and versatile way to handle XML data. However,...Programming Published on 2024-11-18 -
 How Can I Find Users with Today\'s Birthdays Using MySQL?How to Identify Users with Today's Birthdays Using MySQLDetermining if today is a user's birthday using MySQL involves finding all rows where ...Programming Published on 2024-11-18
How Can I Find Users with Today\'s Birthdays Using MySQL?How to Identify Users with Today's Birthdays Using MySQLDetermining if today is a user's birthday using MySQL involves finding all rows where ...Programming Published on 2024-11-18 -
 Using WebSockets in Go for Real-Time CommunicationBuilding apps that require real-time updates—like chat applications, live notifications, or collaborative tools—requires a communication method faster...Programming Published on 2024-11-18
Using WebSockets in Go for Real-Time CommunicationBuilding apps that require real-time updates—like chat applications, live notifications, or collaborative tools—requires a communication method faster...Programming Published on 2024-11-18 -
 Can Class Data Members Be Initialized Directly in C++?Can Class Data Members Be Directly Initialized?In C , class data members cannot be initialized using the direct initialization syntax, (), as seen in...Programming Published on 2024-11-18
Can Class Data Members Be Initialized Directly in C++?Can Class Data Members Be Directly Initialized?In C , class data members cannot be initialized using the direct initialization syntax, (), as seen in...Programming Published on 2024-11-18
Study Chinese
- 1 How do you say "walk" in Chinese? 走路 Chinese pronunciation, 走路 Chinese learning
- 2 How do you say "take a plane" in Chinese? 坐飞机 Chinese pronunciation, 坐飞机 Chinese learning
- 3 How do you say "take a train" in Chinese? 坐火车 Chinese pronunciation, 坐火车 Chinese learning
- 4 How do you say "take a bus" in Chinese? 坐车 Chinese pronunciation, 坐车 Chinese learning
- 5 How to say drive in Chinese? 开车 Chinese pronunciation, 开车 Chinese learning
- 6 How do you say swimming in Chinese? 游泳 Chinese pronunciation, 游泳 Chinese learning
- 7 How do you say ride a bicycle in Chinese? 骑自行车 Chinese pronunciation, 骑自行车 Chinese learning
- 8 How do you say hello in Chinese? 你好Chinese pronunciation, 你好Chinese learning
- 9 How do you say thank you in Chinese? 谢谢Chinese pronunciation, 谢谢Chinese learning
- 10 How to say goodbye in Chinese? 再见Chinese pronunciation, 再见Chinese learning
























